Home › Forums › Main Forum › Experts Corner › Viral Load and SVR › Understanding Viral Load Testing
- This topic has 3 replies, 2 voices, and was last updated 10 years ago by
A.L..
-
AuthorPosts
-
10 January 2016 at 5:00 am #8508
Your will find some information about this here: http://fixhepc.com/forum/experts-corner/364-viral-load-quantitative-vs-qualitative.html
Where I note that:
Hep C Viral Load can be quantitative (undetectable, <15, or 15-100,000,000) or qualitative (undetected/detected).
But what does that look like under the hood?
There is very little viral RNA in your body, so to find it and count it we do this:
- Make an RNA soup out of your blood
- Perform what is known as Polymerase Chain Reaction to amplify the quantity of RNA (rather like brewing beer)
- Have a look at the resulting soup
And here is to sort of thing we see:
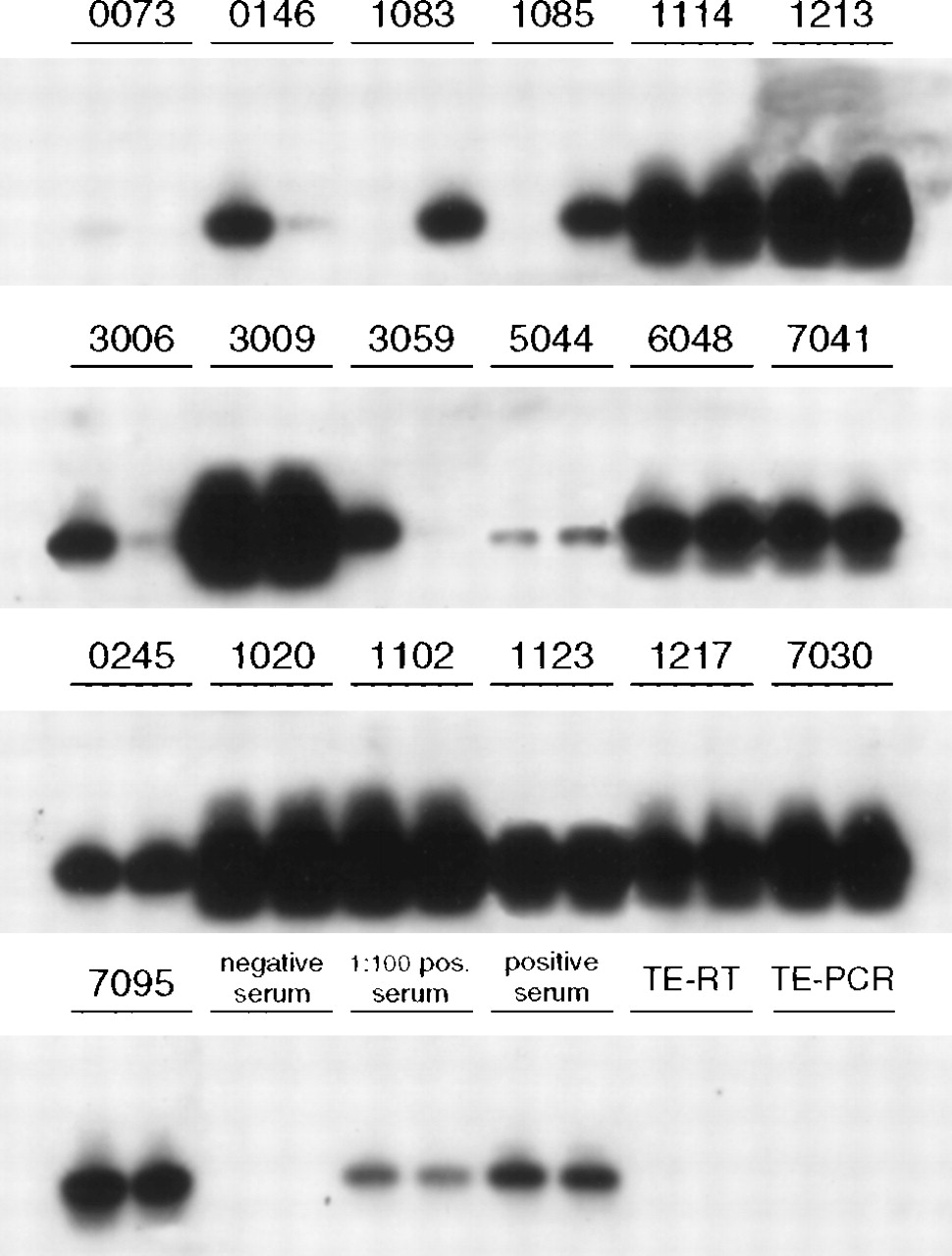
So if you look at the bottom “negative serum” you see we have nothing. With 0073 and 5044 we have faint smudges. 0073 might be reported HCV RNA < 15 (quantitative) HCV RNA detected, and 5044 HCV RNA 42, HCV RNA detected. If you look at the positive serum, to the left you see 1:100 positive serum - this is where the "soup" was diluted 1 part soup to 100 parts water to dilute it - as you can see it still shows up, but is much fainter. Dilution lets us quantify differences between the large solid smudges. When we get to really low levels of RNA all we see really faint smudges. At some point the smudge is there "detected" but to faint to count. The <12, <15, <20 and <25 results we see relate to the minimum sensitivity of the test in use. The lowest we used to be able to get to was <25, but is now typically <15 and recently I have seen <12 in some results from Spain. Although it is obviously great to have nothing at all seen ie <15 and undetected this does not map as well to SVR as you would expect. No, I don't understand that either! http://fixhepc.com/forum/questions-and-answers/541-hcv-rna-undetected.html?start=18
YMMV
10 January 2016 at 6:41 am #8510Very interesting! This is the kind of informative post we don’t often see (anywhere).
Can I ask, please, what is the difference between the 2 traces for each sample. Some of them are not perfect pairs, so I assume there is a difference of some kind.
A.L.
G4, F4, cirrhosis.
Thank you to Gilead, Michael Sofia, and the terrific folk at FixHepC for making this adventure possible.
YEAR….. ALT….. AST….. GGT… FERRITIN………………………………….
2009……. 210….. 215….. 953….. 1400……….. (Bad health, stupidity)
2015……. 60……. 45……. 150….. 360…………. (Improved diet and health, FixHepC treatment)
2016……. 20……. 24……. 25……. 156…………. (SVR 12)10 January 2016 at 8:06 am #8514Can I ask, please, what is the difference between the 2 traces for each sample. Some of them are not perfect pairs, so I assume there is a difference of some kind.
Yes, good pickup.
I chose this image because it had a nice lot of comparator samples all on the one page and they are HCV RNA runs.
These images are Southern Blot HCV RNA comparing before and after seroconversion HCV RNA results.
Here’s the source article which is interesting in and of itself in that it looks at the process of becoming HCV Antibody positive as it compares to serum HCV RNA levels.
http://www.bloodjournal.org/content/94/4/1183?sso-checked=true
YMMV
13 January 2016 at 6:55 am #8895Thanks for that. Interesting attached article (analysing 358 drug users in an HIV study that began in 1985 and then finding 19 of them who became infected with HCV. Just trying to imagine the logistics involved with paperwork and specimen collection and storage with that amount of people, especially considering that the paper itself was published in 1999.)
G4, F4, cirrhosis.
Thank you to Gilead, Michael Sofia, and the terrific folk at FixHepC for making this adventure possible.
YEAR….. ALT….. AST….. GGT… FERRITIN………………………………….
2009……. 210….. 215….. 953….. 1400……….. (Bad health, stupidity)
2015……. 60……. 45……. 150….. 360…………. (Improved diet and health, FixHepC treatment)
2016……. 20……. 24……. 25……. 156…………. (SVR 12) -
AuthorPosts
- You must be logged in to reply to this topic.
